This is my first blog as a Natural Talent Trainee with The Conservation Volunteers, and I’m really excited to document my progress. Already I’m in the 5th week of my placement with SASA (Science and Advice for Scottish Agriculture, part of the Scottish Government), and the time seems to be flying by! My placement this year will be a mixture of learning classical invertebrate ID skills alongside using molecular biology to study their DNA. An opportunity like this, which will teach me both sides of identification is amazing, and I’m looking forward to the year ahead.
The Induction week
Before I started my placement, I attended a week’s induction training with TCV, where all of the Natural Talent and Natural Networks trainees from across the UK met up in Edinburgh. It was great to meet so many new trainees like myself and to hear about their experiences that led up to where they are now. I know that we’ll all definitely keep in touch. We’re already bouncing ideas off each other and seeing where we can work together on different projects.
My favourite part of the week was the day trip to Argyll. Although it didn’t go quite as planned due to a broken down coach…it was still a great day in which we got to enjoy the spectacular Trossachs scenery, and we even got to see some Golden Eagles.
Starting as SASA
My placement at SASA is an entomology traineeship role that will primarily focus on learning how to identify aphids. Aphids are vectors (carriers) of plant viruses, some of which can seriously damage seed potato crops. SASA operates a national network of suction traps collecting information about aphid populations. Through this they can provide advice on the risk of potato virus transmission and the need for aphid control. With Scotland being a major producer of quality seed potatoes, it’s obvious why tracking certain aphid species is so important!
Classical Taxonomy
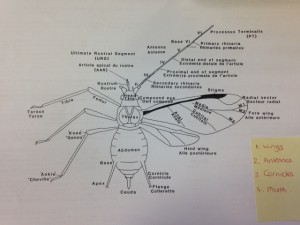
A major part of my traineeship role with SASA will be learning classical taxonomy skills for identifying insects like aphids. Currently, I am a beginner when it comes to aphid ID, so I’m really looking forward to seeing my progress in a year’s time. Last week I started some background reading into aphid morphology and learning the basics about their anatomy and structure. The first stage of my classical ID training was to identify aphids from all other insect species, like in the picture below. It’s pretty incredible looking at these tiny invertebrates down the microscope – there is so much detail that we just can’t see with the naked eye!
Aphids are pretty interesting creatures. What I find most interesting about them is the fact that females can reproduce through parthenogenesis – giving birth to daughter aphids without any fertilisation at all! The daughter aphids can then in turn have offspring by parthenogenesis too. No wonder some aphids are often seen as being pests!
I have also been busy helping to check the vast SASA aphid collections that date all the way back to the 1970s!
Molecular taxonomy
For most of these first few weeks I have been learning about using molecular biology as a tool for identifying species. Small invertebrates can often be extremely difficult to identify, with some species having no clear morphological distinctions between each other. Molecular taxonomy offers an invaluable addition to help identify or to confirm a species by looking at its DNA. I have been learning how to do exactly that in the laboratory. Specifically, I’ve been looking at small insects called psyllids, which are also Hemiptera (true bugs) like aphids. Some species of psyllids can also be responsible for carrying certain crop diseases. Soon, I’ll be applying what I’ve learnt with the psyllids to the aphids. In the lab so far I’ve learnt:
- How to extract the DNA from an insect specimen without destroying the exoskeleton. The voucher specimen can then be kept as an example for that particular species.
- How to run a PCR (Polymerase Chain Reaction), making multiple copies of the DNA.
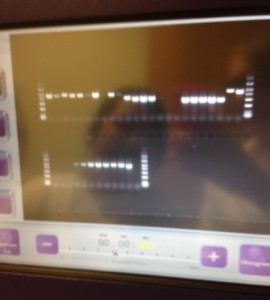
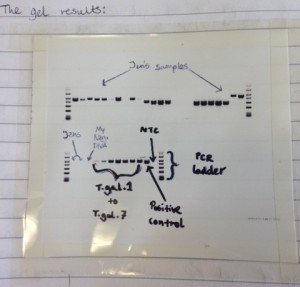
- How to run the DNA through an agarose gel (separating the different sizes of DNA).
- How to run a real-time PCR (which displays the growth of the targeted DNA sequence in real time).
- How to use a variety of other lab equipment e.g. using pipettes, centrifuges, incubators etc.
I’ve not practised some of these lab processes since I was at university, so it’s really good to get to grips with this again. For some others, I’m learning them for the first time. It’s been brilliant so far to be able to shadow the experts, learn and carry out lab work. Being able to see the actual DNA sequence of an insect which I helped to prepare was fascinating.
I can’t believe that it’s been just over a month already. I look forward to sharing more about my traineeship with you all next month!
Rebecca Cairns